|
| https://dire.dcode.org/ |
|
|||||||||
|
DiRE Is Running.... You'll see this screen while DiRE is running. Note the job ID at the top, you can use it later to return to your query. It can take up to 30 minutes for a job to complete, depending on the number of signal and background genes supplied.
Main Output After a successful completion of your query, DiRE will display a results page, divided into four sections as follows: Request ID If you didn't copy this 16-digit number from the running screen, do it now! It will allow you to return to your results without having to run your query again.
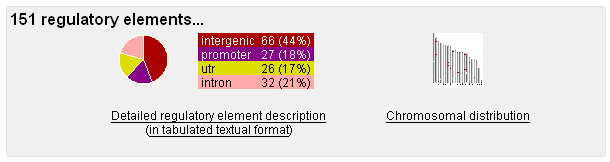
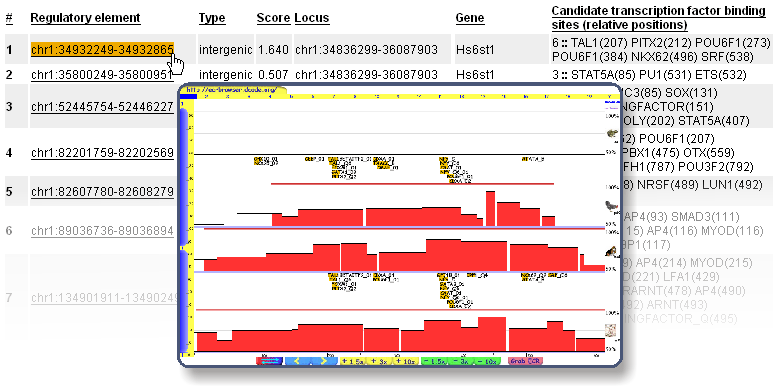
Regulatory Elements In a nutshell, here is what you were looking for!
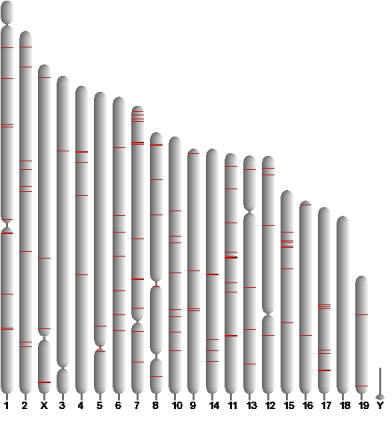
You can quickly see where the predicted regulatory elements are located relative to the genes they presumably regulate, but you can also get a glance of their genomic distribution, indicated by the red bars on the chromosomal representations.
A link to the details is also provided, which will take you on a screen like this:
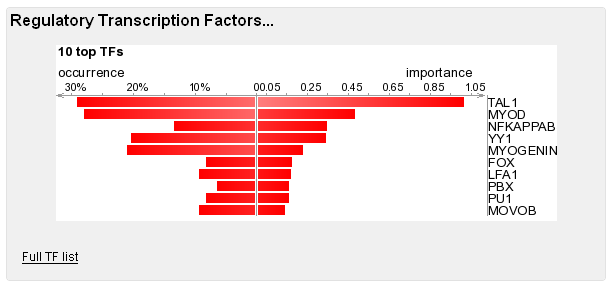
As you can see, you're only one click away from visualizing any regulatory element in the ECR Browser, where you can have a more detailed picture of the locus. Should you need to resort the list, column headers will help you with that. Transcription Factors They make up the regulatory elements, and the 10 most important ones are highlighted in this section.
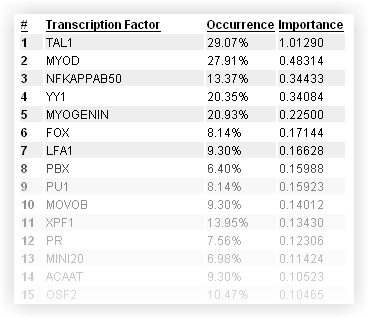
The complete list of TFs that are positively associated with the co-regulated gene is also provided. For each TF you will find its occurrence in REs, and its importance(computed as in Pennacchio et al. 2007). As in the case of REs, you can sort the list with the help of the column headers.
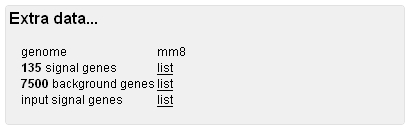
Original Data Back to the roots... For your convenience, original data is available through these links. Note that the original input list, as well as its mapping result onto the genome, is provided. They can contain a different number of entries, because more than once accession number can map to the same gene locus, but it is also possible that one accession number will map to more than one locus. Should you need to run the analysis again with a different set of background genes, you'll find it conveninent to get the original list of genes from here.
Feel free to write to dcode@ncbi.nlm.nih.gov if you have further questions or suggestions. | |||||||||
| NCBI DCODE.org Comparative Genomics Developments (www.dcode.org) | |||||||||